43 KiB
Investigaton of Monte-Carlo Methods
Init
Required Modules
import numpy as np
import matplotlib.pyplot as plt
import monte_carloUtilities
%run ../utility.py
%load_ext autoreload
%aimport monte_carlo
%autoreload 1The autoreload extension is already loaded. To reload it, use: %reload_ext autoreload
Implementation
Center of Mass Frame
"""
Implementation of the analytical cross section for q q_bar ->
gamma gamma
Author: Valentin Boettcher <hiro@protagon.space>
"""
import numpy as np
# NOTE: a more elegant solution would be a decorator
def energy_factor(charge, esp):
"""
Calculates the factor common to all other values in this module
Arguments:
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
return charge ** 4 / (137.036 * esp) ** 2 / 6
def diff_xs(θ, charge, esp):
"""
Calculates the differential cross section as a function of the
azimuth angle θ in units of 1/GeV².
Here dΩ=sinθdθdφ
Arguments:
θ -- azimuth angle
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
f = energy_factor(charge, esp)
return f * ((np.cos(θ) ** 2 + 1) / np.sin(θ) ** 2)
def diff_xs_cosθ(cosθ, charge, esp):
"""
Calculates the differential cross section as a function of the
cosine of the azimuth angle θ in units of 1/GeV².
Here dΩ=d(cosθ)dφ
Arguments:
cosθ -- cosine of the azimuth angle
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
f = energy_factor(charge, esp)
return f * ((cosθ ** 2 + 1) / (1 - cosθ ** 2))
def diff_xs_eta(η, charge, esp):
"""
Calculates the differential cross section as a function of the
pseudo rapidity of the photons in units of 1/GeV^2.
This is actually the crossection dσ/(dφdη).
Arguments:
η -- pseudo rapidity
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
f = energy_factor(charge, esp)
return f * (np.tanh(η) ** 2 + 1)
def diff_xs_p_t(p_t, charge, esp):
"""
Calculates the differential cross section as a function of the
transverse momentum (p_t) of the photons in units of 1/GeV^2.
This is actually the crossection dσ/(dφdp_t).
Arguments:
p_t -- transverse momentum in GeV
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
f = energy_factor(charge, esp)
sqrt_fact = np.sqrt(1 - (2 * p_t / esp) ** 2)
return f / p_t * (1 / sqrt_fact + sqrt_fact)
def total_xs_eta(η, charge, esp):
"""
Calculates the total cross section as a function of the pseudo
rapidity of the photons in units of 1/GeV^2. If the rapditiy is
specified as a tuple, it is interpreted as an interval. Otherwise
the interval [-η, η] will be used.
Arguments:
η -- pseudo rapidity (tuple or number)
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementar charge
"""
f = energy_factor(charge, esp)
if not isinstance(η, tuple):
η = (-η, η)
if len(η) != 2:
raise ValueError("Invalid η cut.")
def F(x):
return np.tanh(x) - 2 * x
return 2 * np.pi * f * (F(η[0]) - F(η[1]))Analytical Integration
And now calculate the cross section in picobarn.
xs_gev = total_xs_eta(η, charge, esp)
xs_pb = gev_to_pb(xs_gev)
tex_value(xs_pb, unit=r'\pico\barn', prefix=r'\sigma = ',
prec=6, save=('results', 'xs.tex'))\(\sigma = \SI{0.053793}{\pico\barn}\)
Lets plot the total xs as a function of η.
fig, ax = set_up_plot()
η_s = np.linspace(0, 3, 1000)
ax.plot(η_s, gev_to_pb(total_xs_eta(η_s, charge, esp)))
ax.set_xlabel(r'$\eta$')
ax.set_ylabel(r'$\sigma$ [pb]')
ax.set_xlim([0, max(η_s)])
ax.set_ylim(0)
save_fig(fig, 'total_xs', 'xs', size=[2.5, 2.5])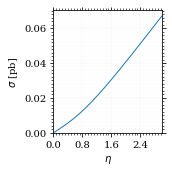
Compared to sherpa, it's pretty close.
sherpa = 0.05380
xs_pb - sherpa-6.7112594623469635e-06
I had to set the runcard option EW_SCHEME: alpha0 to use the pure
QED coupling constant.
Numerical Integration
Plot our nice distribution:
plot_points = np.linspace(*plot_interval, 1000)
fig, ax = set_up_plot()
ax.plot(plot_points, gev_to_pb(diff_xs(plot_points, charge=charge, esp=esp)))
ax.set_xlabel(r'$\theta$')
ax.set_ylabel(r'$d\sigma/d\Omega$ [pb]')
ax.set_xlim([plot_points.min(), plot_points.max()])
ax.axvline(interval[0], color='gray', linestyle='--')
ax.axvline(interval[1], color='gray', linestyle='--', label=rf'$|\eta|={η}$')
ax.legend()
save_fig(fig, 'diff_xs', 'xs', size=[2.5, 2.5])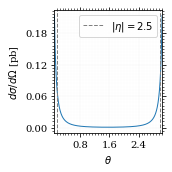
Define the integrand.
def xs_pb_int(θ):
return 2*np.pi*gev_to_pb(np.sin(θ)*diff_xs(θ, charge=charge, esp=esp))
def xs_pb_int_η(η):
return 2*np.pi*gev_to_pb(diff_xs_eta(η, charge, esp))Plot the integrand. # TODO: remove duplication
fig, ax = set_up_plot()
ax.plot(plot_points, xs_pb_int(plot_points))
ax.set_xlabel(r'$\theta$')
ax.set_ylabel(r'$2\pi\cdot d\sigma/d\theta [pb]')
ax.set_xlim([plot_points.min(), plot_points.max()])
ax.axvline(interval[0], color='gray', linestyle='--')
ax.axvline(interval[1], color='gray', linestyle='--', label=rf'$|\eta|={η}$')
save_fig(fig, 'xs_integrand', 'xs', size=[3, 2.2])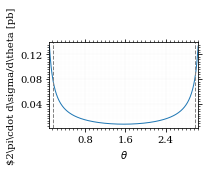
Integral over θ
Intergrate σ with the mc method.
xs_pb_res = monte_carlo.integrate(xs_pb_int, interval, epsilon=1e-3)
xs_pb_resIntegrationResult(result=0.05380076774689264, sigma=0.0009156046547219115, N=2383)
We gonna export that as tex.
tex_value(*xs_pb_res.combined_result, unit=r'\pico\barn',
prefix=r'\sigma = ', save=('results', 'xs_mc.tex'))
tex_value(xs_pb_res.N, prefix=r'N = ', save=('results', 'xs_mc_N.tex'))\(N = 2383\)
Integration over η
Plot the intgrand of the pseudo rap.
fig, ax = set_up_plot()
points = np.linspace(-4, 4, 1000)
ax.set_xlim([-4, 4])
ax.plot(points, xs_pb_int_η(points))
ax.set_xlabel(r'$\eta$')
ax.set_ylabel(r'$2\pi\cdot d\sigma/d\eta$ [pb]')
ax.axvline(interval_η[0], color='gray', linestyle='--')
ax.axvline(interval_η[1], color='gray', linestyle='--', label=rf'$|\eta|={η}$')
save_fig(fig, 'xs_integrand_eta', 'xs', size=[3, 2])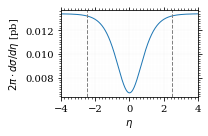
xs_pb_η = monte_carlo.integrate(xs_pb_int_η,
interval_η, epsilon=1e-3)
xs_pb_ηIntegrationResult(result=0.053381188045107726, sigma=0.0009626020740618223, N=137)
As we see, the result is a little better if we use pseudo rapidity, because the differential cross section does not difverge anymore. But becase our η interval is covering the range where all the variance is occuring, the improvement is rather marginal.
And yet again export that as tex.
tex_value(*xs_pb_η.combined_result, unit=r'\pico\barn', prefix=r'\sigma = ',
save=('results', 'xs_mc_eta.tex'))
tex_value(xs_pb_η.N, prefix=r'N = ', save=('results', 'xs_mc_eta_N.tex'))\(N = 137\)
Using VEGAS
Now we use VEGAS on the θ parametrisation and see what happens.
num_increments = 11
xs_pb_vegas = monte_carlo.integrate_vegas(
xs_pb_int,
interval,
num_increments=num_increments,
alpha=1,
increment_epsilon=0.001,
acumulate=False,
)
xs_pb_vegasVegasIntegrationResult(result=0.05353797358506919, sigma=0.00013283395611126086, N=2805, increment_borders=array([0.16380276, 0.2374099 , 0.34568281, 0.5094011 , 0.76958263,
1.23235422, 1.91714873, 2.37499285, 2.63412137, 2.79640826,
2.90423418, 2.9777899 ]), vegas_iterations=9500)
This is pretty good, although the variance reduction may be achieved partially by accumulating the results from all runns. Here this gives us one order of magnitude more than we wanted.
And export that as tex.
tex_value(*xs_pb_vegas.combined_result, unit=r'\pico\barn',
prefix=r'\sigma = ', save=('results', 'xs_mc_θ_vegas.tex'))
tex_value(xs_pb_vegas.N, prefix=r'N = ', save=('results', 'xs_mc_θ_vegas_N.tex'))
tex_value(num_increments, prefix=r'K = ', save=('results', 'xs_mc_θ_vegas_K.tex'))\(K = 11\)
Surprisingly, acumulation, the result ain't much different. This depends, of course, on the iteration count.
monte_carlo.integrate_vegas(
xs_pb_int,
interval,
num_increments=num_increments,
alpha=1,
increment_epsilon=0.001,
acumulate=True,
)VegasIntegrationResult(result=0.053785157752621354, sigma=1.887850854134335e-05, N=2805, increment_borders=array([0.16380276, 0.23716945, 0.34540806, 0.50930035, 0.76937514,
1.22945366, 1.91539567, 2.37452676, 2.63392477, 2.79705411,
2.9048259 , 2.9777899 ]), vegas_iterations=2563)
Let's define some little helpers.
"""
Some shorthands for common plotting tasks related to the investigation
of monte-carlo methods in one rimension.
Author: Valentin Boettcher <hiro at protagon.space>
"""
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import yoda as yo
from collections.abc import Iterable
import yoda.plotting as yplt
import numpy as np
from utility import *
def plot_increments(ax, increment_borders, label=None, *args, **kwargs):
"""Plot the increment borders from a list. The first and last one
:param ax: the axis on which to draw
:param list increment_borders: the borders of the increments
:param str label: the label to apply to one of the vertical lines
"""
ax.axvline(x=increment_borders[1], label=label, *args, **kwargs)
for increment in increment_borders[1:-1]:
ax.axvline(x=increment, *args, **kwargs)
def plot_vegas_weighted_distribution(
ax, points, dist, increment_borders, *args, **kwargs
):
"""Plot the distribution with VEGAS weights applied.
:param ax: axis
:param points: points
:param dist: distribution
:param increment_borders: increment borders
"""
num_increments = increment_borders.size
weighted_dist = dist.copy()
for left_border, right_border in zip(increment_borders[:-1], increment_borders[1:]):
length = right_border - left_border
mask = (left_border <= points) & (points <= right_border)
weighted_dist[mask] = dist[mask] * num_increments * length
ax.plot(points, weighted_dist, *args, **kwargs)
def plot_stratified_rho(ax, points, increment_borders, *args, **kwargs):
"""Plot the weighting distribution resulting from the increment
borders.
:param ax: axis
:param points: points
:param increment_borders: increment borders
"""
num_increments = increment_borders.size
ρ = np.empty_like(points)
for left_border, right_border in zip(increment_borders[:-1], increment_borders[1:]):
length = right_border - left_border
mask = (left_border <= points) & (points <= right_border)
ρ[mask] = 1 / (num_increments * length)
ax.plot(points, ρ, *args, **kwargs)And now we plot the integrand with the incremens.
fig, ax = set_up_plot()
ax.set_xlim(*interval)
ax.set_xlabel(r"$\theta$")
ax.set_ylabel(r"$2\pi\cdot d\sigma/d\theta$ [pb]")
ax.set_ylim([0, 0.09])
ax.plot(plot_points, xs_pb_int(plot_points), label="Distribution")
plot_increments(
ax,
xs_pb_vegas.increment_borders,
label="Increment Borders",
color="gray",
linestyle="--",
)
plot_vegas_weighted_distribution(
ax,
plot_points,
xs_pb_int(plot_points),
xs_pb_vegas.increment_borders,
label="Weighted Distribution",
)
ax.legend(fontsize="small", loc="lower left")
save_fig(fig, "xs_integrand_vegas", "xs", size=[5, 3])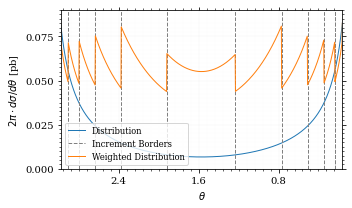
Testing the Statistics
Let's battle test the statistics.
num_runs = 1000
num_within = 0
for _ in range(num_runs):
val, err = \
monte_carlo.integrate(xs_pb_int, interval, epsilon=1e-3).combined_result
if abs(xs_pb - val) <= err:
num_within += 1
num_within/num_runs0.691
So we see: the standard deviation is sound.
Doing the same thing with VEGAS works as well.
num_runs = 1000
num_within = 0
for _ in range(num_runs):
val, err = \
monte_carlo.integrate_vegas(xs_pb_int, interval,
num_increments=10, alpha=1,
epsilon=1e-3, acumulate=False,
vegas_point_density=100).combined_result
if abs(xs_pb - val) <= err:
num_within += 1
num_within/num_runs0.686
Sampling and Analysis
Define the sample number.
sample_num = 1_000_000
tex_value(
sample_num, prefix="N = ", save=("results", "4imp-sample-size.tex"),
)\(N = 1000000\)
Let's define shortcuts for our distributions. The 2π are just there for formal correctnes. Factors do not influecence the outcome.
def dist_cosθ(x):
return gev_to_pb(diff_xs_cosθ(x, charge, esp))
def dist_η(x):
return gev_to_pb(diff_xs_eta(x, charge, esp))Sampling the cosθ cross section
Now we monte-carlo sample our distribution. We observe that the efficiency his very bad!
cosθ_sample, cosθ_efficiency = \
monte_carlo.sample_unweighted_array(sample_num, dist_cosθ,
interval_cosθ, report_efficiency=True,
cache='cache/bare_cos_theta',
proc='auto')
cosθ_efficiency0.02744127583441648
Let's save that.
tex_value(
cosθ_efficiency * 100,
prefix=r"\mathfrak{e} = ",
suffix=r"\%",
save=("results", "naive_th_samp.tex"),
)\(\mathfrak{e} = 3\%\)
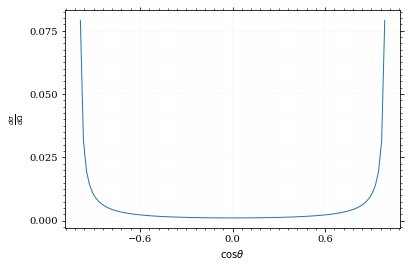
Our distribution has a lot of variance, as can be seen by plotting it.
pts = np.linspace(*interval_cosθ, 100)
fig, ax = set_up_plot()
ax.plot(pts, dist_cosθ(pts))
ax.set_xlabel(r'$\cos\theta$')
ax.set_ylabel(r'$\frac{d\sigma}{d\Omega}$')Text(0, 0.5, '$\\frac{d\\sigma}{d\\Omega}$')
We define a friendly and easy to integrate upper limit function.
fig, ax = set_up_plot()
upper_limit = dist_cosθ(interval_cosθ[0]) / interval_cosθ[0] ** 2
upper_base = dist_cosθ(0)
def upper(x):
return upper_base + upper_limit * x ** 2
def upper_int(x):
return upper_base * x + upper_limit * x ** 3 / 3
ax.plot(pts, upper(pts), label="upper bound")
ax.plot(pts, dist_cosθ(pts), label=r"$f_{\cos\theta}$")
ax.legend(fontsize='small')
ax.set_xlabel(r"$\cos\theta$")
ax.set_ylabel(r"$\frac{d\sigma}{d\cos\theta}$ [pb]")
save_fig(fig, "upper_bound", "xs_sampling", size=(3, 2.5))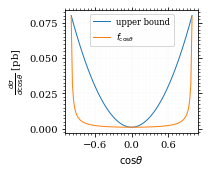
To increase our efficiency, we have to specify an upper bound. That is at least a little bit better. The numeric inversion is horribly inefficent.
cosθ_sample_tuned, cosθ_efficiency_tuned = monte_carlo.sample_unweighted_array(
sample_num,
dist_cosθ,
interval_cosθ,
report_efficiency=True,
proc="auto",
cache="cache/bare_cos_theta_tuned",
upper_bound=[upper, upper_int],
)
cosθ_efficiency_tuned0.07903728511692736
<<cosθ-bare-eff>>
tex_value(
cosθ_efficiency_tuned * 100,
prefix=r"\mathfrak{e} = ",
suffix=r"\%",
save=("results", "tuned_th_samp.tex"),
)\(\mathfrak{e} = 8\%\)
Nice! And now draw some histograms.
We define an auxilliary method for convenience.
def draw_histogram(
ax,
histogram,
errorbars=True,
hist_kwargs=dict(color="#1f77b4"),
errorbar_kwargs=dict(color="orange"),
normalize_to=None,
):
"""Draws a histogram with optional errorbars using the step style.
:param ax: axis to draw on
:param histogram: an array of the form [heights, edges]
:param hist_kwargs: keyword args to pass to `ax.step`
:param errorbar_kwargs: keyword args to pass to `ax.errorbar`
:param normalize_to: if set, the histogram will be normalized to the value
:returns: the given axis
"""
heights, edges = histogram
centers = (edges[1:] + edges[:-1]) / 2
deviations = (
(errorbars if isinstance(errorbars, (np.ndarray, list)) else np.sqrt(heights))
if errorbars is not False
else None
)
if normalize_to is not None:
integral = hist_integral(histogram)
heights = heights / integral * normalize_to
if errorbars is not False:
deviations = deviations / integral * normalize_to
if errorbars is not False:
ax.errorbar(centers, heights, deviations, linestyle="none", **errorbar_kwargs)
ax.step(edges, [heights[0], *heights], **hist_kwargs)
ax.set_xlim(*[edges[0], edges[-1]])
return ax
@functools.wraps(yplt.plot)
def yoda_plot_wrapper(*args, **kwargs):
fig, axs = yplt.plot(*args, **kwargs)
if isinstance(axs, Iterable):
axs = [set_up_axis(ax) for ax in axs if ax is not None]
else:
axs = set_up_axis(axs)
return fig, axs
def samples_to_yoda(samples, bins, range=None, **kwargs):
if range is None:
range = [min(samples), max(samples)]
hist = yo.Histo1D(bins, *range, **kwargs)
for sample in samples:
hist.fill(sample)
return hist
def draw_histo_auto(
points, xlabel, title="", bins=50, range=None, rethist=False, **kwargs
):
"""Creates a histogram figure from sample points, normalized to unity.
:param points: samples
:param xlabel: label of the x axis
:param bins: number of bins
:param range: the range of the values
:param rethist: whether to return the histogram as third argument
:returns: figure, axis
"""
hist = yo.Histo1D(
bins, *(range if range is not None else [min(points), max(points)]), title
)
for point in points:
hist.fill(point)
plot = yoda_plot_wrapper(hist, xlabel=xlabel, **kwargs)
return (*plot, hist) if rethist else plotThe histogram for cosθ.
from importlib import reload
reload(yplt)
fig, _, hist_cosθ = draw_histo_auto(
cosθ_sample,
r"$\cos\theta$",
title=r"own implementation",
rethist=True,
range=interval_cosθ,
)
save_fig(fig, "histo_cos_theta", "xs", size=(4, 3))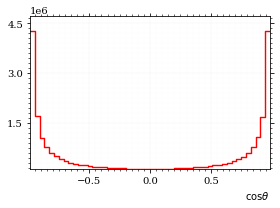
Observables
Now we define some utilities to draw real 4-momentum samples.
@numpy_cache("momentum_cache")
def sample_momenta(sample_num, interval, charge, esp, seed=None, **kwargs):
"""Samples `sample_num` unweighted photon 4-momenta from the
cross-section. Superflous kwargs are passed on to
`sample_unweighted_array`.
:param sample_num: number of samples to take
:param interval: cosθ interval to sample from
:param charge: the charge of the quark
:param esp: center of mass energy
:param seed: the seed for the rng, optional, default is system
time
:returns: an array of 4 photon momenta
:rtype: np.ndarray
"""
cosθ_sample = monte_carlo.sample_unweighted_array(
sample_num, lambda x: diff_xs_cosθ(x, charge, esp), interval_cosθ, **kwargs
)
φ_sample = np.random.uniform(0, 1, sample_num)
def make_momentum(esp, cosθ, φ):
sinθ = np.sqrt(1 - cosθ ** 2)
return np.array([1, sinθ * np.cos(φ), sinθ * np.sin(φ), cosθ],) * esp / 2
momenta = np.array(
[make_momentum(esp, cosθ, φ) for cosθ, φ in np.array([cosθ_sample, φ_sample]).T]
)
return momentaTo generate histograms of other obeservables, we have to define them as functions on 4-impuleses. Using those to transform samples is analogous to transforming the distribution itself.
"""This module defines some observables on arrays of 4-pulses."""
import numpy as np
def p_t(p):
"""Transverse momentum
:param p: array of 4-momenta
"""
return np.linalg.norm(p[:,1:3], axis=1)
def η(p):
"""Pseudo rapidity.
:param p: array of 4-momenta
"""
return np.arccosh(np.linalg.norm(p[:,1:], axis=1)/p_t(p))*np.sign(p[:, 3])And import them.
%aimport tangled.observables
obs = tangled.observablesLets try it out.
momentum_sample = sample_momenta(
sample_num,
interval_cosθ,
charge,
esp,
cache="cache/bare_cos_theta",
proc=3,
momentum_cache="cache/momenta_bare_cos_theta",
)
momentum_sampleTrying cache
array([[100. , 54.98191622, 34.53483728, -76.05480855],
[100. , 87.95719691, 30.53342542, -36.48618154],
[100. , 29.02952127, 15.22393108, -94.47496397],
...,
[100. , 37.62519445, 37.66247038, -84.65153907],
[100. , 75.22515369, 0.79738586, 65.88277793],
[100. , 26.58309333, 4.84203005, -96.28028819]])
Now let's make a histogram of the η distribution.
η_sample = obs.η(momentum_sample)
fig, ax, hist_obs_η = draw_histo_auto(
η_sample, r"$\eta$", title="own implementation", range=interval_η, rethist=True
)
hist_obs_η.normalize(1)
save_fig(fig, "histo_eta", "xs_sampling", size=[3, 3])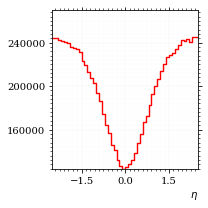
And the same for the p_t (transverse momentum) distribution.
p_t_sample = obs.p_t(momentum_sample)
fig, ax, hist_obs_pt = draw_histo_auto(
p_t_sample,
r"$p_T$ [GeV]",
title="own implementation",
range=interval_pt,
rethist=True,
)
save_fig(fig, "histo_pt", "xs_sampling", size=[3, 3]) BrokenPipeErrorTraceback (most recent call last)
<ipython-input-202-d07958ed2945> in <module>
7 rethist=True,
8 )
----> 9 save_fig(fig, "histo_pt", "xs_sampling", size=[3, 3])
~/Documents/Projects/UNI/Bachelor/prog/python/qqgg/utility.py in save_fig(fig, title, folder, size)
189
190 fig.savefig(f"./figs/{folder}/{title}.pdf")
--> 191 fig.savefig(f"./figs/{folder}/{title}.pgf")
192
193
/usr/lib/python3.8/site-packages/matplotlib/figure.py in savefig(self, fname, transparent, **kwargs)
2201 self.patch.set_visible(frameon)
2202
-> 2203 self.canvas.print_figure(fname, **kwargs)
2204
2205 if frameon:
/usr/lib/python3.8/site-packages/matplotlib/backend_bases.py in print_figure(self, filename, dpi, facecolor, edgecolor, orientation, format, bbox_inches, **kwargs)
2096
2097 try:
-> 2098 result = print_method(
2099 filename,
2100 dpi=dpi,
/usr/lib/python3.8/site-packages/matplotlib/backends/backend_pgf.py in print_pgf(self, fname_or_fh, *args, **kwargs)
888 if not cbook.file_requires_unicode(file):
889 file = codecs.getwriter("utf-8")(file)
--> 890 self._print_pgf_to_fh(file, *args, **kwargs)
891
892 def _print_pdf_to_fh(self, fh, *args, **kwargs):
/usr/lib/python3.8/site-packages/matplotlib/cbook/deprecation.py in wrapper(*args, **kwargs)
356 f"%(removal)s. If any parameter follows {name!r}, they "
357 f"should be pass as keyword, not positionally.")
--> 358 return func(*args, **kwargs)
359
360 return wrapper
/usr/lib/python3.8/site-packages/matplotlib/backends/backend_pgf.py in _print_pgf_to_fh(self, fh, dryrun, bbox_inches_restore, *args, **kwargs)
870 RendererPgf(self.figure, fh),
871 bbox_inches_restore=bbox_inches_restore)
--> 872 self.figure.draw(renderer)
873
874 # end the pgfpicture environment
/usr/lib/python3.8/site-packages/matplotlib/artist.py in draw_wrapper(artist, renderer, *args, **kwargs)
36 renderer.start_filter()
37
---> 38 return draw(artist, renderer, *args, **kwargs)
39 finally:
40 if artist.get_agg_filter() is not None:
/usr/lib/python3.8/site-packages/matplotlib/figure.py in draw(self, renderer)
1733
1734 self.patch.draw(renderer)
-> 1735 mimage._draw_list_compositing_images(
1736 renderer, self, artists, self.suppressComposite)
1737
/usr/lib/python3.8/site-packages/matplotlib/image.py in _draw_list_compositing_images(renderer, parent, artists, suppress_composite)
135 if not_composite or not has_images:
136 for a in artists:
--> 137 a.draw(renderer)
138 else:
139 # Composite any adjacent images together
/usr/lib/python3.8/site-packages/matplotlib/artist.py in draw_wrapper(artist, renderer, *args, **kwargs)
36 renderer.start_filter()
37
---> 38 return draw(artist, renderer, *args, **kwargs)
39 finally:
40 if artist.get_agg_filter() is not None:
/usr/lib/python3.8/site-packages/matplotlib/axes/_base.py in draw(self, renderer, inframe)
2628 renderer.stop_rasterizing()
2629
-> 2630 mimage._draw_list_compositing_images(renderer, self, artists)
2631
2632 renderer.close_group('axes')
/usr/lib/python3.8/site-packages/matplotlib/image.py in _draw_list_compositing_images(renderer, parent, artists, suppress_composite)
135 if not_composite or not has_images:
136 for a in artists:
--> 137 a.draw(renderer)
138 else:
139 # Composite any adjacent images together
/usr/lib/python3.8/site-packages/matplotlib/artist.py in draw_wrapper(artist, renderer, *args, **kwargs)
36 renderer.start_filter()
37
---> 38 return draw(artist, renderer, *args, **kwargs)
39 finally:
40 if artist.get_agg_filter() is not None:
/usr/lib/python3.8/site-packages/matplotlib/axis.py in draw(self, renderer, *args, **kwargs)
1239 self._update_label_position(renderer)
1240
-> 1241 self.label.draw(renderer)
1242
1243 self._update_offset_text_position(ticklabelBoxes, ticklabelBoxes2)
/usr/lib/python3.8/site-packages/matplotlib/artist.py in draw_wrapper(artist, renderer, *args, **kwargs)
36 renderer.start_filter()
37
---> 38 return draw(artist, renderer, *args, **kwargs)
39 finally:
40 if artist.get_agg_filter() is not None:
/usr/lib/python3.8/site-packages/matplotlib/text.py in draw(self, renderer)
683
684 with _wrap_text(self) as textobj:
--> 685 bbox, info, descent = textobj._get_layout(renderer)
686 trans = textobj.get_transform()
687
/usr/lib/python3.8/site-packages/matplotlib/text.py in _get_layout(self, renderer)
297 clean_line, ismath = self._preprocess_math(line)
298 if clean_line:
--> 299 w, h, d = renderer.get_text_width_height_descent(
300 clean_line, self._fontproperties, ismath=ismath)
301 else:
/usr/lib/python3.8/site-packages/matplotlib/backends/backend_pgf.py in get_text_width_height_descent(self, s, prop, ismath)
755
756 # get text metrics in units of latex pt, convert to display units
--> 757 w, h, d = (LatexManager._get_cached_or_new()
758 .get_width_height_descent(s, prop))
759 # TODO: this should be latex_pt_to_in instead of mpl_pt_to_in
/usr/lib/python3.8/site-packages/matplotlib/backends/backend_pgf.py in get_width_height_descent(self, text, prop)
356
357 # send textbox to LaTeX and wait for prompt
--> 358 self._stdin_writeln(textbox)
359 try:
360 self._expect_prompt()
/usr/lib/python3.8/site-packages/matplotlib/backends/backend_pgf.py in _stdin_writeln(self, s)
257 self.latex_stdin_utf8.write(s)
258 self.latex_stdin_utf8.write("\n")
--> 259 self.latex_stdin_utf8.flush()
260
261 def _expect(self, s):
BrokenPipeError: [Errno 32] Broken pipe
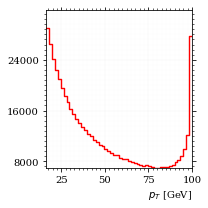
That looks somewhat fishy, but it isn't.
fig, ax = set_up_plot()
points = np.linspace(interval_pt[0], interval_pt[1] - .01, 1000)
ax.plot(points, gev_to_pb(diff_xs_p_t(points, charge, esp)))
ax.set_xlabel(r'$p_T$')
ax.set_xlim(interval_pt[0], interval_pt[1] + 1)
ax.set_ylim([0, gev_to_pb(diff_xs_p_t(interval_pt[1] -.01, charge, esp))])
ax.set_ylabel(r'$\frac{d\sigma}{dp_t}$ [pb]')
save_fig(fig, 'diff_xs_p_t', 'xs_sampling', size=[4, 2])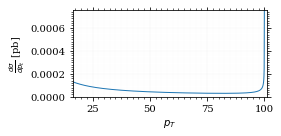 this is strongly peaked at p_t=100GeV. (The jacobian goes like 1/x there!)
this is strongly peaked at p_t=100GeV. (The jacobian goes like 1/x there!)
Sampling the η cross section
An again we see that the efficiency is way, way! better…
η_sample, η_efficiency = monte_carlo.sample_unweighted_array(
1000,
dist_η,
interval_η,
report_efficiency=True,
proc="auto",
#cache="cache/sample_bare_eta",
)
tex_value(
η_efficiency * 100,
prefix=r"\mathfrak{e} = ",
suffix=r"\%",
save=("results", "eta_eff.tex"),
)\(\mathfrak{e} = 41\%\)
<<η-eff>>
Let's draw a histogram to compare with the previous results.
η_hist = samples_to_yoda(η_sample, 50, title="sampled from $d\sigma / d\eta$")
reload(yplt)
η_hist.normalize(1)
η_hist.setAnnotation('ratioref', True)
yoda_plot_wrapper([hist_obs_η, η_hist], errorbars=True)
# fig, (ax_hist, ax_ratio) = draw_ratio_plot(
# [
# dict(hist=η_hist, hist_kwargs=dict(label=r"sampled from $d\sigma / d\eta$"),),
# dict(
# hist=hist_obs_η,
# hist_kwargs=dict(
# label=r"sampled from $d\sigma / d\cos\theta$", color="black"
# ),
# ),
# ],
# )
# ax_hist.legend(loc="upper center", fontsize="small")
# ax_ratio.set_xlabel(r"$\eta$")
#save_fig(fig, "comparison_eta", "xs_sampling", size=(4, 4))| <Figure | size | 576x432 | with | 2 | Axes> | (<matplotlib.axes._subplots.AxesSubplot at 0x7f923b1ea070> <matplotlib.axes._subplots.AxesSubplot at 0x7f923b18bdc0>) |
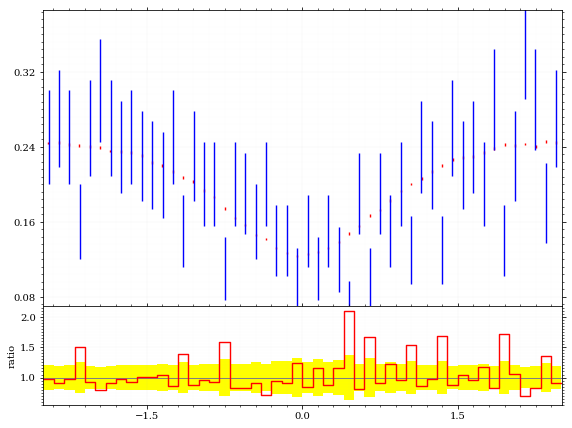
Looks good to me :).
Sampling with VEGAS
To get the increments, we have to let VEGAS loose on our
distribution. We throw away the integral, but keep the increments.
K = 10
increments = monte_carlo.integrate_vegas(
dist_cosθ, interval_cosθ, num_increments=K, alpha=1, increment_epsilon=0.001
).increment_borders
tex_value(
K, prefix=r"K = ", save=("results", "vegas_samp_num_increments.tex"),
)
incrementsarray([-0.9866143 , -0.96980073, -0.93072072, -0.83891589, -0.6017223 ,
0.00829172, 0.6071626 , 0.83981042, 0.93107467, 0.96951636,
0.9866143 ])
Visualizing the increment borders gives us the information we want.
pts = np.linspace(*interval_cosθ, 100)
fig, ax = set_up_plot()
ax.plot(pts, dist_cosθ(pts))
ax.set_xlabel(r'$\cos\theta$')
ax.set_ylabel(r'$\frac{d\sigma}{d\Omega}$')
ax.set_xlim(*interval_cosθ)
plot_increments(ax, increments,
label='Increment Borderds', color='gray', linestyle='--')
ax.legend()<matplotlib.legend.Legend at 0x7f924d64e730>
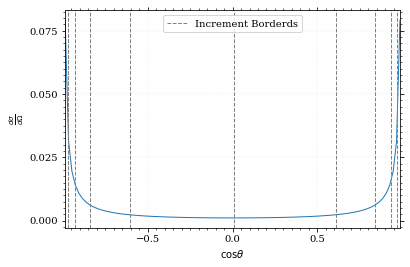
We can now plot the reweighted distribution to observe the variance reduction visually.
pts = np.linspace(*interval_cosθ, 1000)
fig, ax = set_up_plot()
ax.plot(pts, dist_cosθ(pts), label="Distribution")
plot_vegas_weighted_distribution(
ax, pts, dist_cosθ(pts), increments, label="Weighted Distribution"
)
ax.set_xlabel(r"$\cos\theta$")
ax.set_ylabel(r"$\frac{d\sigma}{d\cos\theta}$")
ax.set_xlim(*interval_cosθ)
plot_increments(
ax, increments, label="Increment Borderds", color="gray", linestyle="--"
)
ax.legend(fontsize="small")
save_fig(fig, "vegas_strat_dist", "xs_sampling", size=(3, 2.3))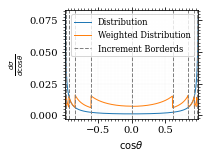
I am batman! Let's plot the weighting distribution.
pts = np.linspace(*interval_cosθ, 1000)
fig, ax = set_up_plot()
plot_stratified_rho(ax, pts, increments)
ax.set_xlabel(r"$\cos\theta$")
ax.set_ylabel(r"$\rho")
ax.set_xlim(*interval_cosθ)
save_fig(fig, "vegas_rho", "xs_sampling", size=(3, 2.3))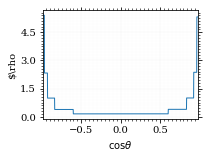
Now, draw a sample and look at the efficiency.
cosθ_sample_strat, cosθ_efficiency_strat = monte_carlo.sample_unweighted_array(
sample_num,
dist_cosθ,
increment_borders=increments,
report_efficiency=True,
proc="auto",
cache="cache/sample_bare_cos_theta_vegas",
)
cosθ_efficiency_strat0.5954571428571428
tex_value(
cosθ_efficiency_strat * 100,
prefix=r"\mathfrak{e} = ",
suffix=r"\%",
save=("results", "strat_th_samp.tex"),
)\(\mathfrak{e} = 60\%\)
If we compare that to /hiro/bachelor_thesis/src/commit/f157b525328f012d5d5ea1acc6820047e5af6f55/prog/python/qqgg/cos%CE%B8-bare-eff, we can see the improvement :P. It is even better the /hiro/bachelor_thesis/src/commit/f157b525328f012d5d5ea1acc6820047e5af6f55/prog/python/qqgg/%CE%B7-eff. The histogram looks just the same.
fig, _ = draw_histo_auto(cosθ_sample_strat, r'$\cos\theta$')
save_fig(fig, 'histo_cos_theta_strat', 'xs', size=(4,3)) TypeErrorTraceback (most recent call last)
<ipython-input-148-ff4d6a24ae42> in <module>
----> 1 fig, _ = draw_histo_auto(cosθ_sample_strat, r'$\cos\theta$')
2 save_fig(fig, 'histo_cos_theta_strat', 'xs', size=(4,3))
<ipython-input-132-507ea080dcd6> in draw_histo_auto(points, xlabel, bins, range, rethist, **kwargs)
70
71
---> 72 return yoda_nplot_wrapper(hist, xlabel=xlabel, **kwargs)
<ipython-input-132-507ea080dcd6> in yoda_nplot_wrapper(*args, **kwargs)
42 @functools.wraps(yplt.nplot)
43 def yoda_nplot_wrapper(*args, **kwargs):
---> 44 fig, axs = yplt.nplot(*args, **kwargs)
45 if isinstance(axs, iterable):
46 axs = [set_up_axis(ax) for ax in axs]
/usr/lib/python3.8/site-packages/yoda/plotting.py in nplot(hs, outfiles, ratio, show, nproc, **plotkeys)
446 else:
447 ## Run this way in the 1 proc case for easier debugging
--> 448 rtn = [_plot1arg(args) for args in argslist]
449
450 if show:
/usr/lib/python3.8/site-packages/yoda/plotting.py in <listcomp>(.0)
446 else:
447 ## Run this way in the 1 proc case for easier debugging
--> 448 rtn = [_plot1arg(args) for args in argslist]
449
450 if show:
/usr/lib/python3.8/site-packages/yoda/plotting.py in _plot1arg(args)
397 def _plot1arg(args):
398 "Helper function for mplot, until Py >= 3.3 multiprocessing.pool.starmap() is available"
--> 399 print(args)
400 return plot(*args)
401
include/generated/Bin1D_Dbn1D.pyx in yoda.core.Bin1D_Dbn1D.__repr__()
TypeError: must be real number, not builtin_function_or_method
Some Histograms with Rivet
Init
import yodaPlot the Histos
def yoda_to_numpy(histo):
histo.normalize(
histo.numEntries() * ((histo.xMax() - histo.xMin()) / histo.numBins())
)
edges = np.append(histo.xMins(), histo.xMax())
heights = histo.yVals().astype(int)
return heights, edges
def draw_yoda_histo_auto(h, xlabel, **kwargs):
hist = yoda_to_numpy(h)
fig, ax = set_up_plot()
draw_histogram(ax, hist, errorbars=True, normalize_to=1, **kwargs)
ax.set_xlabel(xlabel)
return fig, ax yoda_file = yoda.read("../../runcards/qqgg/analysis/Analysis.yoda")
sherpa_histos = {
"pT": dict(reference=hist_obs_pt, label="$p_T$ [GeV]"),
"eta": dict(reference=hist_obs_η, label=r"$\eta$"),
"cos_theta": dict(reference=hist_cosθ, label=r"$\cos\theta$"),
}
for key, sherpa_hist in sherpa_histos.items():
yoda_hist = yoda_to_numpy(yoda_file["/MC_DIPHOTON_SIMPLE/" + key])
label = sherpa_hist["label"]
fig, (ax_hist, ax_ratio) = draw_ratio_plot(
[
dict(
hist=yoda_hist,
hist_kwargs=dict(
label="Sherpa Reference"
),
),
dict(
hist=sherpa_hist["reference"],
hist_kwargs=dict(label="Own Implementation"),
),
],
)
ax_ratio.set_xlabel(label)
save_fig(fig, "histo_sherpa_" + key, "xs_sampling", size=(4, 3.5))
NameErrorTraceback (most recent call last)
<ipython-input-151-c0fdcee231b0> in <module>
1 yoda_file = yoda.read("../../runcards/qqgg/analysis/Analysis.yoda")
2 sherpa_histos = {
----> 3 "pT": dict(reference=hist_obs_pt, label="$p_T$ [GeV]"),
4 "eta": dict(reference=hist_obs_η, label=r"$\eta$"),
5 "cos_theta": dict(reference=hist_cosθ, label=r"$\cos\theta$"),
NameError: name 'hist_obs_pt' is not defined
%aimport yoda.plotting
hist = yoda_file["/MC_DIPHOTON_SIMPLE/pT"]
hist.setAnnotation('ratioref', True)
hist.setAnnotation('Title', "LOL")
#hist.annotationsDict()
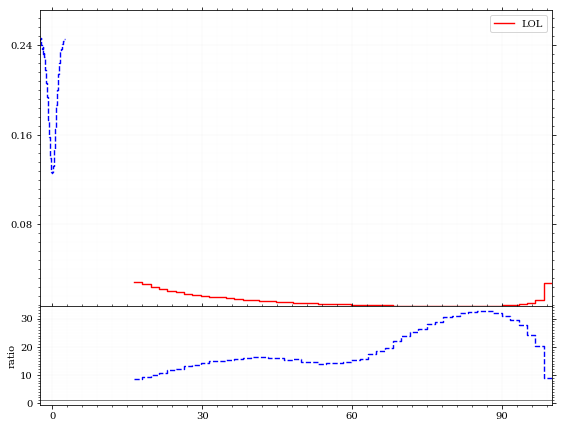
fig, (ax_hist, ax_ratio) =yoda.plotting.plot([yoda_file["/MC_DIPHOTON_SIMPLE/pT"], yoda_file["/MC_DIPHOTON_SIMPLE/eta"]], ratio=True)
set_up_axis(ax_hist)
set_up_axis(ax_ratio)
ax_hist.legend()<matplotlib.legend.Legend at 0x7f924b270910>