19 KiB
Init
Required Modules
import numpy as np
import matplotlib.pyplot as plt
import monte_carloUtilities
%run ../utility.py
%load_ext autoreload
%aimport monte_carlo
%autoreload 1Implementation
"""
Implementation of the analytical cross section for q q_bar ->
gamma gamma
Author: Valentin Boettcher <hiro@protagon.space>
"""
import numpy as np
# NOTE: a more elegant solution would be a decorator
def energy_factor(charge, esp):
"""
Calculates the factor common to all other values in this module
Arguments:
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
return charge**4/(137.036*esp)**2/6
def diff_xs(θ, charge, esp):
"""
Calculates the differential cross section as a function of the
azimuth angle θ in units of 1/GeV².
Here dΩ=sinθdθdφ
Arguments:
θ -- azimuth angle
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
f = energy_factor(charge, esp)
return f*((np.cos(θ)**2+1)/np.sin(θ)**2)
def diff_xs_cosθ(cosθ, charge, esp):
"""
Calculates the differential cross section as a function of the
cosine of the azimuth angle θ in units of 1/GeV².
Here dΩ=d(cosθ)dφ
Arguments:
cosθ -- cosine of the azimuth angle
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
f = energy_factor(charge, esp)
return f*((cosθ**2+1)/(1-cosθ**2))
def diff_xs_eta(η, charge, esp):
"""
Calculates the differential cross section as a function of the
pseudo rapidity of the photons in units of 1/GeV^2.
This is actually the crossection dσ/(dφdη).
Arguments:
η -- pseudo rapidity
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
f = energy_factor(charge, esp)
return f*(np.tanh(η)**2 + 1)
def diff_xs_p_t(p_t, charge, esp):
"""
Calculates the differential cross section as a function of the
transverse momentum (p_t) of the photons in units of 1/GeV^2.
This is actually the crossection dσ/(dφdp_t).
Arguments:
p_t -- transverse momentum in GeV
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementary charge
"""
f = energy_factor(charge, esp)
sqrt_fact = np.sqrt(1-(2*p_t/esp)**2)
return f/p_t*(1/sqrt_fact + sqrt_fact)
def total_xs_eta(η, charge, esp):
"""
Calculates the total cross section as a function of the pseudo
rapidity of the photons in units of 1/GeV^2. If the rapditiy is
specified as a tuple, it is interpreted as an interval. Otherwise
the interval [-η, η] will be used.
Arguments:
η -- pseudo rapidity (tuple or number)
esp -- center of momentum energy in GeV
charge -- charge of the particle in units of the elementar charge
"""
f = energy_factor(charge, esp)
if not isinstance(η, tuple):
η = (-η, η)
if len(η) != 2:
raise ValueError('Invalid η cut.')
def F(x):
return np.tanh(x) - 2*x
return 2*np.pi*f*(F(η[0]) - F(η[1]))Calculations
First, set up the input parameters.
η = 2.5
charge = 1/3
esp = 200 # GeVSet up the integration and plot intervals.
interval_η = [-η, η]
interval = η_to_θ([-η, η])
interval_cosθ = np.cos(interval)
interval_pt = np.sort(η_to_pt([0, η], esp/2))
plot_interval = [0.1, np.pi-.1]Analytical Integration
And now calculate the cross section in picobarn.
xs_gev = total_xs_eta(η, charge, esp)
xs_pb = gev_to_pb(xs_gev)
tex_value(xs_pb, unit=r'\pico\barn', prefix=r'\sigma = ',
prec=6, save=('results', 'xs.tex'))\(\sigma = \SI{0.053793}{\pico\barn}\)
Lets plot the total xs as a function of η.
fig, ax = set_up_plot()
η_s = np.linspace(0, 3, 1000)
ax.plot(η_s, gev_to_pb(total_xs_eta(η_s, charge, esp)))
ax.set_xlabel(r'$\eta$')
ax.set_ylabel(r'$\sigma$ [pb]')
ax.set_xlim([0, max(η_s)])
ax.set_ylim(0)
save_fig(fig, 'total_xs', 'xs', size=[2.5, 2])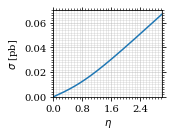
Compared to sherpa, it's pretty close.
sherpa = 0.05380
xs_pb - sherpa-6.7112594623469635e-06
I had to set the runcard option EW_SCHEME: alpha0 to use the pure
QED coupling constant.
Numerical Integration
Plot our nice distribution:
plot_points = np.linspace(*plot_interval, 1000)
fig, ax = set_up_plot()
ax.plot(plot_points, gev_to_pb(diff_xs(plot_points, charge=charge, esp=esp)))
ax.set_xlabel(r'$\theta$')
ax.set_ylabel(r'$d\sigma/d\Omega$ [pb]')
ax.axvline(interval[0], color='gray', linestyle='--')
ax.axvline(interval[1], color='gray', linestyle='--', label=rf'$|\eta|={η}$')
ax.legend()
save_fig(fig, 'diff_xs', 'xs', size=[2.5, 2])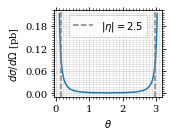
Define the integrand.
def xs_pb_int(θ):
return 2*np.pi*gev_to_pb(np.sin(θ)*diff_xs(θ, charge=charge, esp=esp))
def xs_pb_int_η(η):
return 2*np.pi*gev_to_pb(diff_xs_eta(η, charge, esp))Plot the integrand. # TODO: remove duplication
fig, ax = set_up_plot()
ax.plot(plot_points, xs_pb_int(plot_points))
ax.set_xlabel(r'$\theta$')
ax.set_ylabel(r'$\sin(\theta)\cdot\frac{d\sigma}{d\Omega}$ [pb]')
ax.axvline(interval[0], color='gray', linestyle='--')
ax.axvline(interval[1], color='gray', linestyle='--', label=rf'$|\eta|={η}$')
ax.legend()
save_fig(fig, 'xs_integrand', 'xs', size=[4, 4])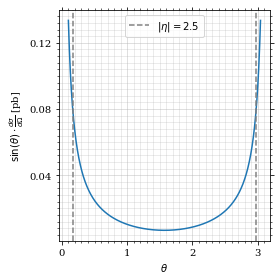
Integral over θ
Intergrate σ with the mc method.
xs_pb_mc, xs_pb_mc_err = monte_carlo.integrate(xs_pb_int, interval, 1000)
xs_pb_mc = xs_pb_mc
xs_pb_mc, xs_pb_mc_err| 0.05323177940348952 | 0.000836179760412404 |
We gonna export that as tex.
tex_value(xs_pb_mc, unit=r'\pico\barn', prefix=r'\sigma = ', err=xs_pb_mc_err, save=('results', 'xs_mc.tex'))\(\sigma = \SI{0.0543\pm 0.0008}{\pico\barn}\)
Integration over η
Plot the intgrand of the pseudo rap.
fig, ax = set_up_plot()
points = np.linspace(*interval_η, 1000)
ax.plot(points, xs_pb_int_η(points))
ax.set_xlabel(r'$\eta$')
ax.set_ylabel(r'$\frac{d\sigma}{d\theta}$ [pb]')
save_fig(fig, 'xs_integrand_η', 'xs', size=[4, 4])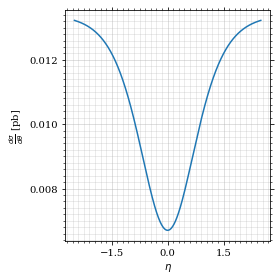
As we see, the result is much better if we use pseudo rapidity, because the differential cross section does not difverge anymore.
xs_pb_η = monte_carlo.integrate(xs_pb_int_η,
interval_η, 1000)
xs_pb_η| 0.05369352543075011 | 0.0001566582384086374 |
And yet again export that as tex.
tex_value(*xs_pb_η, unit=r'\pico\barn', prefix=r'\sigma = ', save=('results', 'xs_mc_eta.tex'))\(\sigma = \SI{0.05398\pm 0.00016}{\pico\barn}\)
Using VEGAS
Now we use VEGAS on the θ parametrisation and see what happens.
xs_pb_vegas, xs_pb_vegas_σ, xs_θ_intervals = \
monte_carlo.integrate_vegas(xs_pb_int, interval,
num_increments=20, alpha=4,
point_density=1000, acumulate=True)
xs_pb_vegas, xs_pb_vegas_σshtsh
| 0.053806254940947366 | 5.91849792512895e-05 |
This is pretty good, although the variance reduction may be achieved partially by accumulating the results from all runns. The uncertainty is being overestimated!
And export that as tex.
tex_value(xs_pb_vegas, xs_pb_vegas_σ, unit=r'\pico\barn',
prefix=r'\sigma = ', save=('results', 'xs_mc_θ_vegas.tex'))\(\sigma = \SI{0.05383\pm 0.00007}{\pico\barn}\)
Surprisingly, without acumulation, the result ain't much different. This depends, of course, on the iteration count.
monte_carlo.integrate_vegas(xs_pb_int, interval, num_increments=20,
alpha=4, point_density=1000,
acumulate=False)[0:2]| 0.05386167571815434 | 7.519896920354165e-05 |
Testing the Statistics
Let's battle test the statistics.
num_runs = 1000
num_within = 0
for _ in range(num_runs):
val, err = monte_carlo.integrate(xs_pb_int_η, interval_η, 1000)
if abs(xs_pb - val) <= err:
num_within += 1
num_within/num_runs0.671
So we see: the standard deviation is sound.
Doing the same thing with VEGAS shows, that we overestimate σ here.
num_runs = 1000
num_within = 0
for _ in range(num_runs):
val, err, _ = \
monte_carlo.integrate_vegas(xs_pb_int, interval,
num_increments=20, alpha=4,
point_density=1000, acumulate=False)
if abs(xs_pb - val) <= err:
num_within += 1
num_within/num_runs0.727
Sampling and Analysis
Define the sample number.
sample_num = 1000Let's define shortcuts for our distributions. The 2π are just there for formal correctnes. Factors do not influecence the outcome.
def dist_θ(x):
return gev_to_pb(diff_xs_cosθ(x, charge, esp))*2*np.pi
def dist_η(x):
return gev_to_pb(diff_xs_eta(x, charge, esp))*2*np.piSampling the cosθ cross section
Now we monte-carlo sample our distribution. We observe that the efficiency his very bad!
cosθ_sample, cosθ_efficiency = \
monte_carlo.sample_unweighted_array(sample_num, dist_θ,
interval_cosθ, report_efficiency=True)
cosθ_efficiency0.026983702912102593
Our distribution has a lot of variance, as can be seen by plotting it.
pts = np.linspace(*interval_cosθ, 100)
fig, ax = set_up_plot()
ax.plot(pts, dist_θ(pts), label=r'$\frac{d\sigma}{d\Omega}$')| <matplotlib.lines.Line2D | at | 0x7f71fa2accd0> |
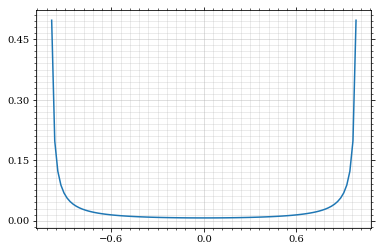
We define a friendly and easy to integrate upper limit function.
upper_limit = dist_θ(interval_cosθ[0]) \
/interval_cosθ[0]**2
upper_base = dist_θ(0)
def upper(x):
return upper_base + upper_limit*x**2
def upper_int(x):
return upper_base*x + upper_limit*x**3/3
ax.plot(pts, upper(pts), label='Upper bound')
ax.legend()
ax.set_xlabel(r'$\cos\theta$')
ax.set_ylabel(r'$\frac{d\sigma}{d\Omega}$')
save_fig(fig, 'upper_bound', 'xs_sampling', size=(4, 4))
fig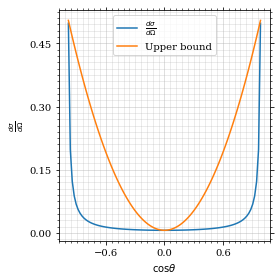
To increase our efficiency, we have to specify an upper bound. That is at least a little bit better. The numeric inversion is horribly inefficent.
cosθ_sample, cosθ_efficiency = \
monte_carlo.sample_unweighted_array(sample_num, dist_θ,
interval_cosθ, report_efficiency=True,
upper_bound=[upper, upper_int])
cosθ_efficiency0.08121827411167512
Nice! And now draw some histograms.
We define an auxilliary method for convenience.
import matplotlib.pyplot as plt
def draw_histo(points, xlabel, bins=20):
heights, edges = np.histogram(points, bins)
centers = (edges[1:] + edges[:-1])/2
deviations = np.sqrt(heights)
fig, ax = set_up_plot()
ax.errorbar(centers, heights, deviations, linestyle='none', color='orange')
ax.step(edges, [heights[0], *heights], color='#1f77b4')
ax.set_xlabel(xlabel)
ax.set_xlim([points.min(), points.max()])
return fig, axThe histogram for cosθ.
fig, _ = draw_histo(cosθ_sample, r'$\cos\theta$')
save_fig(fig, 'histo_cos_theta', 'xs', size=(4,3))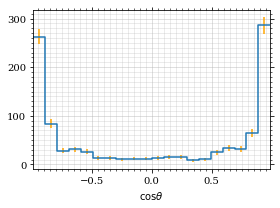
Observables
Now we define some utilities to draw real 4-momentum samples.
def sample_momenta(sample_num, interval, charge, esp, seed=None):
"""Samples `sample_num` unweighted photon 4-momenta from the
cross-section.
:param sample_num: number of samples to take
:param interval: cosθ interval to sample from
:param charge: the charge of the quark
:param esp: center of mass energy
:param seed: the seed for the rng, optional, default is system
time
:returns: an array of 4 photon momenta
:rtype: np.ndarray
"""
cosθ_sample = \
monte_carlo.sample_unweighted_array(sample_num,
lambda x:
diff_xs_cosθ(x, charge, esp),
interval_cosθ)
φ_sample = np.random.uniform(0, 1, sample_num)
def make_momentum(esp, cosθ, φ):
sinθ = np.sqrt(1-cosθ**2)
return np.array([1, sinθ*np.cos(φ), sinθ*np.sin(φ), cosθ])*esp/2
momenta = np.array([make_momentum(esp, cosθ, φ) \
for cosθ, φ in np.array([cosθ_sample, φ_sample]).T])
return momentaTo generate histograms of other obeservables, we have to define them as functions on 4-impuleses. Using those to transform samples is analogous to transforming the distribution itself.
"""This module defines some observables on arrays of 4-pulses."""
import numpy as np
def p_t(p):
"""Transverse momentum
:param p: array of 4-momenta
"""
return np.linalg.norm(p[:,1:3], axis=1)
def η(p):
"""Pseudo rapidity.
:param p: array of 4-momenta
"""
return np.arccosh(np.linalg.norm(p[:,1:], axis=1)/p_t(p))*np.sign(p[:, 3])Lets try it out.
momentum_sample = sample_momenta(2000, interval_cosθ, charge, esp)
momentum_samplearray([[100. , 14.99955553, 6.52933179, -98.65283149],
[100. , 48.11160501, 71.52596373, -50.68836134],
[100. , 27.36251906, 1.55938536, -96.17099806],
...,
[100. , 98.44690501, 13.80044529, 10.85147935],
[100. , 17.20635886, 4.27420589, 98.41581366],
[100. , 66.84034758, 32.63142055, 66.83979599]])
Now let's make a histogram of the η distribution.
η_sample = η(momentum_sample)
draw_histo(η_sample, r'$\eta$')| <Figure | size | 432x288 | with | 1 | Axes> | <matplotlib.axes._subplots.AxesSubplot | at | 0x7f71fa804310> |
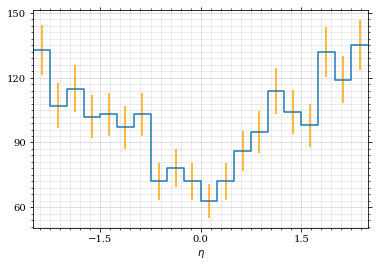
And the same for the p_t (transverse momentum) distribution.
p_t_sample = p_t(momentum_sample)
draw_histo(p_t_sample, r'$p_T$ [GeV]')| <Figure | size | 432x288 | with | 1 | Axes> | <matplotlib.axes._subplots.AxesSubplot | at | 0x7f71fa8a4250> |
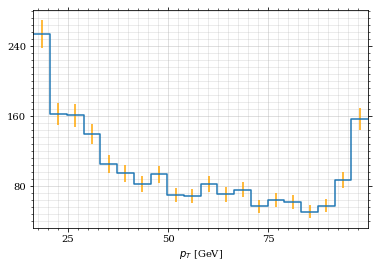
That looks somewhat fishy, but it isn't.
fig, ax = set_up_plot()
points = np.linspace(interval_pt[0], interval_pt[1] - .01, 1000)
ax.plot(points, gev_to_pb(diff_xs_p_t(points, charge, esp)))
ax.set_xlabel(r'$p_T$')
ax.set_xlim(interval_pt[0], interval_pt[1] + 1)
ax.set_ylim([0, gev_to_pb(diff_xs_p_t(interval_pt[1] -.01, charge, esp))])
ax.set_ylabel(r'$\frac{d\sigma}{dp_t}$ [pb]')
save_fig(fig, 'diff_xs_p_t', 'xs_sampling', size=[4, 3])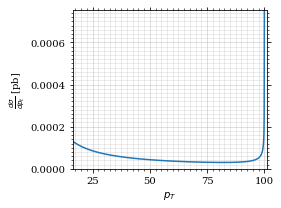 this is strongly peaked at p_t=100GeV. (The jacobian goes like 1/x there!)
this is strongly peaked at p_t=100GeV. (The jacobian goes like 1/x there!)
Sampling the η cross section
An again we see that the efficiency is way, way! better…
η_sample, η_efficiency = \
monte_carlo.sample_unweighted_array(sample_num, dist_η,
interval_η, report_efficiency=True)
η_efficiency0.3973333333333333
Let's draw a histogram to compare with the previous results.
draw_histo(η_sample, r'$\eta$')| <Figure | size | 432x288 | with | 1 | Axes> | <matplotlib.axes._subplots.AxesSubplot | at | 0x7f71fa59e820> |
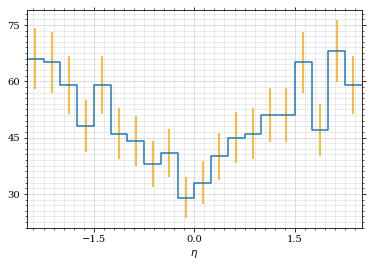
Looks good to me :).